Factorial Arrangement Feedlot Beef Steers Additives
Abstract
Diets for feedlot cattle must be a higher energy density, entailing high fermentable carbohydrate content. Feed additives are needed to reduce possible metabolic disorders. This study aimed to analyze the post-rumen effects of different levels of starch (25%, 35%, and 45%) and additives (monensin or a blend of essential oils and exogenous α-amylase) in diets for Nellore feedlot cattle. The cecum tissue proteome was analyzed via two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) and then differentially expressed protein spots were identified with liquid chromatography–tandem mass spectrometry (LC–MS/MS). The use of blends of essential oils associated with α-amylase as a feed additive promoted the upregulation of enzymes such as triosephosphate isomerase, phosphoglycerate mutase, alpha-enolase, beta-enolase, fructose-bisphosphate aldolase, pyruvate kinase, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), l-lactate dehydrogenase B, l-lactate dehydrogenase A chain, l-lactate dehydrogenase, and ATP synthase subunit beta, which promote the degradation of carbohydrates in the glycolysis and gluconeogenesis pathways and oxidative phosphorylation, support pyruvate metabolism through the synthesis of lactate from pyruvate, and participate in the electron transport chain, producing ATP from ADP in the presence of a proton gradient across the membrane. The absence of proteins related to inflammation processes (leukocyte elastase inhibitors) in the cecum tissues of animals fed essential oils and amylase may be because feed enzymes can remain active in the intestine and aid in the digestion of nutrients that escape rumen fermentation; conversely, the effect of monensin is more evident in the rumen and less than 10% results in post-ruminal action, corroborating the hypothesis that ionophore antibiotics have a limited effect on the microbiota and intestinal fermentation of ruminants. However, the increase in starch in these diets promoted a downregulation of enzymes linked to carbohydrate degradation, probably caused by damage to the cecum epithelium due to increased responses linked to inflammatory injuries.
Introduction
Among the limitations to enhancing meat production is its large energy requirement, which means that feedlot cattle have a higher net energy demand (NE)1. Dietary strategies are adopted to increase energy metabolism, mainly through the fermentation of carbohydrates in the rumen. This is mostly accomplished by supplying hexoses (glucose) from starch. Thus, with increased starch in the diet, physiological limits to animals' ability to digest a large amount of fermentable carbohydrates in the rumen and ruminal escape increased.
In the rumen, the fermentation of glucose from starch occurs and it is converted mainly into volatile fatty acids (AGV) and lactate2, which are metabolized in the liver and provide the greatest source of energy for ruminants3,4. However, the use of large amounts of starch can lead to disorders and metabolic diseases due to the accumulation of organic acids in the ruminal fluid, especially acidosis and bloat (NASCEM5). Thus, feed additives that decrease harmful ruminal fermentation processes are employed, such as sodium monensin, which is a polyester carboxylic ionophore used in growth and finishing diets5. It acts bacteriostatically on Gram-positive ruminal bacteria but may leave residues in products of animal origin and result in microbial resistance6. Alternative additives have shown the potential to replace monensin, such as blends of essential oils associated with the exogenous enzyme α-amylase, which has led to demonstrable gains in performance and carcass weight, in addition to reducing hepatic abscesses and fecal starch in animals with high-starch diets7,8,9.
With high levels of starch in the diet, the rate of passage and post-ruminal digestion increases5. The rumen microbiota can digest around 70–80% of the starch consumed5,10,11,12,13; however, the digestion and absorption of post-rumen starch are partially impaired as enzymatic digestion via pancreatic α-amylase in the duodenum is limited to the small intestine14,15. Others have postulated that glucose cannot be absorbed and transported in large quantities from the lumen into the bloodstream due to insufficient levels of the glucose transporters SLGT1 and GLUT25,16,17, which favors the escape of some starch to the large intestine and increases the potential for the digestion and use of this starch in the cecum. Therefore, feedlot diets that offer increased amounts of energy due to high levels of concentrates18 can cause excessive fermentation in the cecum, contributing to the metabolizable energy ruminants can access19,20 but potentially resulting in hindgut acidosis, which may generate inflammatory reactions in the cecal epithelium. Large amounts of starch in the cecum may contribute to the fermentation of AGV, NH3, and lactic acid as well as a decreased pH5. Additionally, the cecum has limited buffering capacity compared with the rumen, where saliva and protozoa modulate pH fluctuations21. Feed additives that can increase the use of starch in the rumen, reducing starch escape to the intestines, as well as lower starch levels in feedlot cattle diets can avert the risk of cecal acidification. Most studies with feed additives focus only on the rumen effects; however, the effects of feed additives on the post-rumen digestive tract are important to understand.
Understanding how the digestion and absorption sites act when high proportions of starch are included in feedlot diets is essential. Due to the levels of starch in cattle diets and the effects on the extent of the gastrointestinal tract associated with different feed additives, this study aims to map the proteome of feedlot cattle's ceca and to elucidate how protein expression acts on metabolism when different nutritional strategies are applied.
Results
Image analysis and protein expression
In the workspace, classes were created to analyze differences in protein expression; an analysis of variance (ANOVA) was used to test the hypothesis (Hθ) that the expressed spots are identical. When testing all classes, protein spots were differentially expressed, as shown in Table 1.
Supplemental Figure S2 shows the distribution of proteins and their biological processes, molecular functions, and cellular components.
Proteins associated with glucose metabolism and energy synthesis (Table 2) and macromolecules involved in the degradation of carbohydrates through the glycolytic pathway, gluconeogenesis, and oxidative phosphorylation were detected in cecal tissue. The expression of seven enzymes participating in the glycolysis and gluconeogenesis pathways was verified: triosephosphate isomerase (Step 1); phosphoglycerate mutase (Step 2); alpha-enolase (ENO1), beta-enolase (ENO3), and fructose-bisphosphate aldolase (ALDOB) (Step 4); pyruvate kinase (PKM) (Step 5); and glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Three enzymes linked to pyruvate metabolism or catalytic activities participating in the synthesis of lactate from pyruvate were verified as well: l-lactate dehydrogenase B, l-lactate dehydrogenase A chain, and l-lactate dehydrogenase. ATP synthase subunit beta participated in the electron transport chain, producing ATP from ADP in the presence of a proton gradient across the membrane.
Pathways enrichment and Reactome analysis
The pathway enrichment and Reactome analysis yielded similar results showing that specific pathways were affected. The differential expression found in all groups displayed changes in metabolic pathways as carbohydrate metabolism, pyruvate metabolism, the citric acid (TCA) cycle, respiratory electron transport, innate immune system, and the immune system were affected in cecum tissues by different feeding strategies (Fig. 1). The data from the Reactome pathway analysis have been provided in Supplemental Table S1.
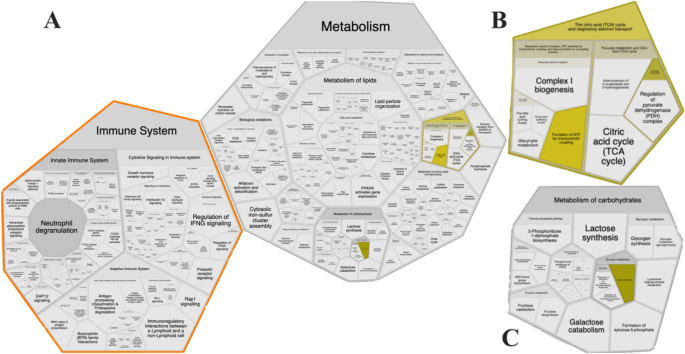
Affected pathways generated from KEGG ID input using Reactome show that immune system and metabolism is impacted (A), which, glycolysis and gluconeogenesis (B) and metabolism of carbohydrates (C).
Additionally, the differential expression indicates similar encoding enzymes in the glycolysis and gluconeogenesis pathways in cattle's large intestines under different feeding strategies (Fig. 2).
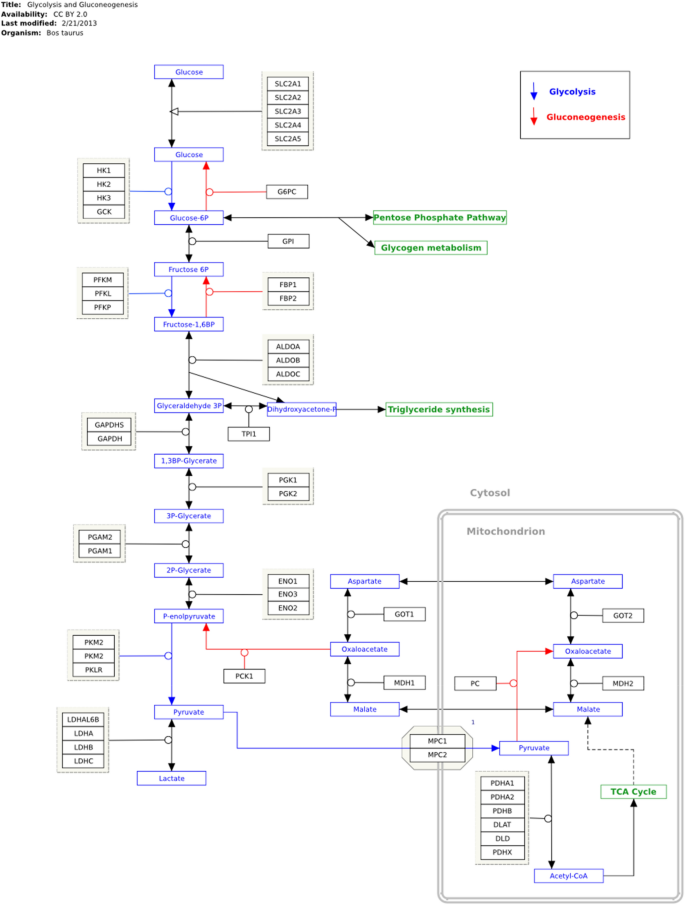
Expression protein profile encoding enzymes in glycolysis and gluconeogenesis pathway. KEGG key: EC 4.1.2.13: Fructose-bisphosphate aldolase (ALDOB); EC 5.3.1.1: Triosephosphate isomerase (TPI); EC 1.2.1.12: Glyceraldehyde-3-phosphate dehydrogenase (GAPDH); EC 5.4.2.4: Phosphoglycerate mutase (PGAM); Alpha-enolase (ENO1); EC 4.2.1.11 Beta-enolase (ENO3); EC 2.7.1.40 Pyruvate Kinase (PKM); EC 1.1.1.27 L-lactate dehydrogenase (LDH).
The expression values (\(P \le 0.05\)) (Table 3) were grouped with hierarchical cluster analysis (Fig. 3) and ordered by homogeneity between the treatments tested. Animals fed with identical levels of starch but subjected to different feed additives showed differentiation in proteins that contribute to energy metabolism.
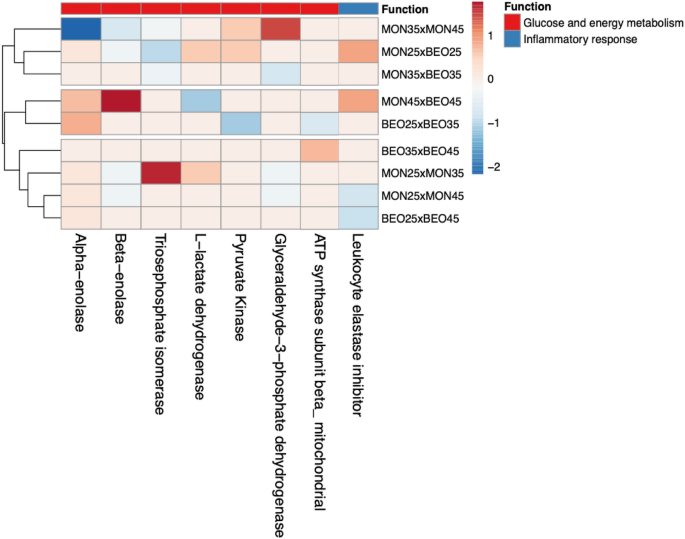
Heatmap of the differentially expressed proteins (ANOVA, P \(\le\) 0.05) among the diets contending different starch levels and additives. Color-coded matrix showed the correlation coefficient of the spots expression values. Each row and column represent one group and protein, respective.
The animals fed low-starch diets (25%) associated with BEOα exhibited increased expression of pyruvate kinase (EC 2.7.1.40), beta-enolase (EC 4.2.1.11), triosephosphate isomerase (EC 5.3.1.1), and l-lactate dehydrogenase (EC 1.1.1.27) compared with those treated with monensin; all of these proteins are enzymes catalyzing the synthesis of pyruvate, which is responsible for the degradation of carbohydrates. Animals fed diets with the highest level of starch tested (45%) exhibited greater synthesis of l-lactate dehydrogenase (EC 1.1.1.27), fructose-bisphosphate aldolase (EC 4.1.2.13), and phosphoglycerate mutase (EC 5.4.2.4). Meanwhile, animals fed the intermediate starch level exhibited a higher expression of triosephosphate isomerase (EC 5.3.1.1) and glyceraldehyde-3-phosphate dehydrogenase (EC 1.2.1.12).
Discussion
When evaluating the tested additives, we found that the use of BEOα with intermediate levels of starch (35%) resulted in greater expression of glycolysis intermediates, thus, this additive may have a greater effect on the post-rumen tract. A companion study (data under review) reported that the optimum level of dietary starch for cattle fed MON was 25%; however, the optimum level of dietary starch for cattle receiving BEO was 35%, commensurate with protein expression synthesis. In summary, feed intake decreased when MON- and BEO-fed cattle were fed more than 25% and 35% starch, respectively, which resulted in decreased average daily gain and proteins linked with carbohydrate degradation. The diets with 45% starch may have caused excessive ruminal fermentation that may have resulted in increased inflammation of the ruminal epithelium, agreeing with the proteins leukocyte elastase inhibitor. The literature reports that increasing levels of starch play an important role in reducing feed intake (observed in a companion study); however, this effect was more evident in cattle that were fed MON as it is a feed additive that depresses intake.
In the protein spots of groups 25BEOα and 45BEOα compared with those fed MON, leukocyte elastase inhibitor, a serine protease inhibitor that is essential for the regulation of inflammatory responses and limits the activity of inflammatory caspases, was not expressed22 aligning with the results of the above-mentioned authors, who reported reduced ionophore effects in the hindgut. When comparing diets with 25% or 45% starch, regardless of the additive used, leukocyte elastase inhibitor was expressed more, corroborating previous studies demonstrating that inflammatory injuries are caused by the increased use of concentrates in the diet23,24. Additionally, we observed reduced expression of proteins that participate in energy metabolism in animals on high-starch diets, which can damage the epithelium of the cecum.
In a similar study, Toseti et al.9 observed a reduction in fecal starch using BEOα, suggesting a greater degradation of carbohydrates because the feed enzymes can remain active in the intestine and aid in the digestion of nutrients that escape rumen fermentation25. As Thomas et al.26 demonstrated, the effect of monensin is more evident in the rumen, mainly in the diversity of microorganisms, but less than 10% results in post-ruminal action, corroborating the hypothesis that ionophore antibiotics have a limited effect on ruminants' microbiota and intestinal fermentation.
Protein expression differs depending on the dietary starch level (\(P \le 0.05\)); the cluster analysis shows differentiation in the profile of the identified proteins involved in energy metabolism (Fig. 3) but the effect is greater when contrasting 25% and 35% starch or 35% and 45%, mainly when using monensin as a feed additive. In summary, high concentrations of starch may result in inflammatory responses due to the greater supply of undegradable starch in the rumen, thus decreasing the expression of proteins linked to the glycolytic pathway through tissue damage and inflammation. Higher concentrations of carbohydrates (starch) in the intestine along with the low effects of monensin on the cecum may contribute to a greater accumulation of organic acids. Additionally, the intestinal epithelium is much more vulnerable to pH variation than the rumen21,27, corroborating our identification of proteins linked to immune responses. Notably, this was not observed when assessing the full range of starch levels (25% vs. 45%) but the proteins involved in inflammatory responses were expressed more (Fig. 3). We attribute this to the greater increase in dietary carbohydrate, which may have increased epithelial injury (indicative damage)21,28 and upregulated inflammatory response, subsequently reducing the expression of proteins associated with energy metabolism.
Fructose-bisphosphate aldolase (ALDOB), an enzyme that converts fructose-1,6-bisphosphate to fructose 6-phosphate, catalyzed by triosephosphate isomerase (TPI), is a precursor of glyceraldehyde-3-phosphate (GA3P), which is acted upon by the enzyme glyceraldehyde-3-phosphate dehydrogenase (GAPDH) during glycolysis. Alpha-enolase (ENO1) and beta-enolase (ENO3) are isoforms of enolase that are involved in Step 4 of glycolytic metabolism. Phosphoglycerate mutase (PGM) is a catalytic enzyme that converts 3-phosphoglycerate to 2-phosphoglycerate and, finally, pyruvate kinase (PKM) synthesizes pyruvate in the last step of glycolysis (UniProt29). In ruminants, a high concentration of starch enables the fermentation of carbohydrates in the cecum with lactate production, which increases glucose metabolism in the intestine and leads to the observed expression of the enzyme l-lactate dehydrogenase and its isoforms l-lactate dehydrogenase B and l-lactate dehydrogenase A, which synthesize lactate from pyruvate (UniProt29).
The dietary manipulation verified the expression of the leukocyte elastase inhibitor protein, associated with the inflammatory response (Table 2); this plays an essential role in regulating the innate immune response, inflammation, and cellular homeostasis, and mainly acts to protect cell proteases released into the cytoplasm during stress or infection29.
Methods
The experiment was conducted according to the standards issued by the National Council for Animal Experimentation Control (CONCEA) and approved by the Ethics and Use of Animals Committee of the São Paulo State University (UNESP, Botucatu-SP), under protocol no 0107/2019 and in compliance with ARRIVE (animal research: reporting of in vivo experiments) guidelines30.
Animals, facilities, feeding and animal care
The animal experiment was conducted at the feedlot facilities of the Innovation and Applied Science Center of DSM Nutritional Products (I & AS Beef Center; Rio Brilhante, Mato Grosso do Sul, Brazil). Nellore bulls (n = 210) (Bos taurus indicus) from the grazing system with an average body weight of ± 380 kg were used. The animals were randomly allocated to pens (7 animals/pen) with 12 m2/animal and collective troughs (50 cm linear/animal). The program for receiving the animals consisted of weighing, deworming, and vaccinating according to the annual prophylactic calendar. The animals underwent a pre-experimental adaptation period of 10 days to standardize their rumen population and allow them to adapt to the facilities and management. The diets were formulated with the LRNS system (large ruminant nutrition system31), level 2, meeting the nutritional requirements for daily weight gain between 1.5 and 1.7 kg/animal. Animals were fed for 92 days and diets were offered ad libitum twice daily at 8 a. m. and 3 p.m.
Experimental design
A factorial 3 × 2 arrangement was used, with starch level (25%, 35%, or 45%) and additives (monensin or the essential oil blend CRINA® with the exogenous α-amylase Rumistar®) as the factor. The sodium monensin (MON; Rumensin, Elanco Animal Health, Indianapolis, IN) used was included in the diet at a dose of 26 mg/kg of dry matter. The blend of functional oils (CRINA RUMINANTS®; DSM Nutritional Products, Basel, Switzerland) containing thymol, eugenol, limonene, and vanillin32 and the exogenous enzyme α-amylase (RONOZYME RUMISTAR™; DSM Nutritional Products, Basel, Switzerland), referred to as BEOα were added to the diet at a dose of 90 mg/kg of dry matter and 560 mg/kg of dry matter, respectively. The pens were distributed in a randomized block design, totaling 6 treatments with 5 repetitions or 30 experimental units overall. The treatments were distributed within the blocks as follows: T1 (25MON), T2 (25BEOα), T3 (35MON), T4 (35BEOα), T5 (45MON), and T6 (45BEOα). According to the statistical model:
$$Y_{ijk} = \mu + B_{k} + C_{i} + A_{J} + \left( {C \times A} \right)_{ij} + \varepsilon_{ijk} ,$$
where \(Yijk\) is the dependent variable; \(\mu\) is the overall mean; \(Bk\) is the block effect; \(Ci\) is concentrate; \(AJ\) is additive; \((C \times A)ij\) is the interaction between concentrate and additive effects; and \(\varepsilon ijk\) is the residual error.
Diets and their chemical composition
The experimental diets were composed of natural bagasse sugarcane, ground corn, soybean hulls, cottonseed, soybeans, core minerals and vitamins, urea, and additives. The transition to the finishing diet was managed as follows: for 14 days, two diets with 65% and 75% concentrate were provided for 7 days each. From the 15th day of the experiment until slaughter, a finishing diet containing 85% concentrate was provided (Table 4).
Dietary energy content was calculated according to the LRNS system31 and Total digestible nutrients (TDN) were determined by the equation: TDN = digestible CP + (digestible EE × 2.25) + digestible NDF + digestible non-structural carbohydrate (NSC). Crude protein was determined by assessing the nitrogen content of the samples with the Kjeldahl method33. The NDF concentration was assessed with the methodology described by Van Soest et al.34 and corrected for CP and ashes. Starch was determined by the equation: NSC = 100 − CP − EE − NDF − ash, where ash content was determined by incinerating samples at 550 °C for 2 h in a muffle furnace35. Physically effective neutral detergent fiber (peNFD) was determined according to Kononoff et al.'s methods36. Samples of diets were collected to determine particle-size distribution by sieving with the Penn State particle-size separator and reported on an as-fed basis.
Proteomics sample collection and preparation
The animals were transported to a commercial slaughterhouse where they were stunned by brain concussions with a captive dart gun. After bleeding hide removal and evisceration, cecum samples about 4 cm square were collected and washed with phosphate-buffered saline (PBS), transferred to 15 mL polypropylene bottles, and placed in liquid nitrogen (− 196 °C) for later protein extraction. Each pen was considered an experimental unit, so a pool of samples was made by homogenizing cecal tissue from animals given the same treatment; three animals per experimental unit (N = 5) were used, i.e., 15 animals/group or 90 animals total (15 animals from each of six groups).
Extraction, precipitation and quantification of proteins
To extract the protein fraction, the tissue was macerated with a mortar and pestle in the presence of liquid nitrogen. The extracting solution was added at a rate of 1 mL ultrapure water per 1 g tissue and then the samples were homogenized with an OMMI-BEAD RUPTOR4 cell disruptor (Kennesaw, Georgia, United States) over three 30-s cycles. The samples were then separated into protein extracts and the supernatant was collected after refrigerated centrifugation (− 4 °C) with a UNIVERSAL 320R HETTICH (Tuttlingen, Baden-Württemberg, Germany). The proteins were precipitated in 80% (v/v) acetone (J.T. Baker, Phillipsburg, New Jersey, United States), using 300 μL of supernatant and 600 μL of 80% acetone. The samples were stored at 2 °C for 1.5 h and then centrifuged at 14,000 rpm for 30 min; the supernatant was discarded and the protein pellet was solubilized in 1 mL of 0.50 mol/L NaOH (Merck, Darmstadt, Germany). The protein concentrations were determined by the Biuret method37, using an analytical curve with a concentration range of 0–100 g/L of standard bovine albumin solution (Acros Organics, NJ, United States) at 100 g/L.
Electrophoretic separations of protein fractions using 2D-PAGE
For isoelectric focusing, about 375 µg of proteins from each group were applied to individual strips; the sample was resolubilized with a solution containing 7 mol/L urea, 2 mol/L thiourea, 2% CHAPS (m/v) (GE Healthcare, Uppsala, Sweden), ampholytes at a pH of 3 to 10 at 0.5% (v/v) (GE Healthcare, Uppsala, Sweden), and 0.002% bromophenol blue (GE Healthcare, Uppsala, Sweden), in addition to 2.8 mg of dithiothreitol (USB, Cleveland, Ohio, United States). Approximately 900 µL of mineral oil was added at room temperature for 12 h to rehydrate the strips and prevent evaporation and urea crystals. After this period, the strips were added to the EttanTMIPGphorTM3 isoelectric focusing system (IEF) (GE Healthcare, Uppsala, Sweden). The electrical voltage used was established by the protocol described by Braga et al.38. At the end of the focusing period, the strip was balanced in two 15-min stages. First, 10 mL of a solution containing 6 mol/L urea, 2% SDS (w/v), 30% glycerol (v/v), 50 mmol/L Tris–HCl (pH 8.8), 0.002% bromophenol blue (w/v), and 2% DTT (w/v) was used to keep the proteins in their reduced forms38,37. In the second stage, a solution in which DTT was replaced with 2.5% (w/v) iodoacetamide was used to alkylate the thiol groups of the proteins and prevent possible reoxidation. After strip balancing, the second portion of the electrophoretic process (SDS-PAGE) occurred.
The strip was applied to a 12.5% (w/v) polyacrylamide gel previously prepared on a glass plate (180 × 160 × 1.5 mm). The gel was placed next to the strip with a piece of filter paper containing 6 µL of a molecular mass standard (GE Healthcare, Uppsala, Sweden), with proteins of different molecular masses (β-phosphorylase [97.0 kDa], albumin [66.0 kDa], ovalbumin [45.0 kDa], carbonic anhydrase [30.0 kDa], trypsin inhibitor [20.1 kDa], and α-lactalbumin [14.4 kDa]). The strip and filter paper were sealed with 0.5% agarose solution (w/v) to ensure contact with the polyacrylamide gel. The run program was then applied at 100 V for 30 min and a further 250 V for 2 h. After the run period, the gels were immersed in a fixative solution containing10% acetic acid (v/v) and 40% ethanol (v/v) for 30 min. Then, the proteins were revealed with colloidal Coomassie G-250 (USB, Cleveland, Ohio, United States) for 72 h and removed by washing with ultrapure water38,39,40,41.
The gels obtained (Supplemental Fig. S1) were scanned and their images analyzed with the image processing program ImageMaster 2D Platinum 7.0 (GeneBio, Geneva, Switzerland; www.gelifescience.com), which allows the estimation of the isoelectric points and molecular masses of the separated proteins and calculation of the number of spots obtained via gel electrophoresis. Three replicates of each gel were used to evaluate the reproducibility of each protein spot obtained in the replicates of the gels by overlaying the image from one gel over the other in the image treatment program39,40,41,42.
Protein identification by mass spectrometry (LC–MS/MS)
The differentially expressed spots were characterized via mass spectrometry after the identification was standardized according to the highest protein score, pI, and molecular mass (MM) closest to the theoretical and experimental results. Among the proteins identified, 12 were classified as functional for this study as they are related to energy metabolism and inflammatory responses.
The protein spots were characterized with LC–MS/MS after being subjected to tryptic digestion and peptide elution according to the methodology Shevchenko et al.43 described. The aliquots of the solutions containing the eluted peptides were analyzed to obtain the mass spectra with the nanoAcquity UPLC system coupled to the Xevo G2 QTof mass spectrometer (Waters, Milford, MA, United States). Proteins were identified by searching in the UniProt database (www.uniprot.org) within the Bos taurus species. Proteins were considered depending on their theoretical and experimental isoelectric points, molecular masses, and scores (> 60). After identifying FASTA sequences in the proteins, their sequences were analyzed with OMICSBOX software (BLAST2GO)44 and they were categorized by their molecular function, biological processes, and biochemical activities with gene ontology (GO).
Proteomic statistical analysis
The starch level and additive were the fixed effects analyzed in a factorial design; thus, the groups were compared through contrasts to verify differentially expressed protein spots. Only proteins with significantly altered levels were selected for identification by MS. The images were analyzed with ImageMaster Platinum software version 7.0, which establishes correlations (matching) between groups. For this correlation, three gel replicates were compared for volume, distribution, relative intensity, isoelectric point, and molecular mass in an analysis of variance (ANOVA) with a t-test to determine the significance of differentially expressed protein spots.
Following the average mode of background subtraction, individual spot intensity volume was normalized with total intensity volume (the summation of the intensity volumes obtained from all spots in the same 2-DE gel). The normalized intensity volume values of individual protein spots were then used to determine differential protein expression among experimental groups. A heatmap showed the correlation coefficient of the spot expression values and, after checking the differentially expressed spots (t-test, P < 0.05), the log2 FC values were used for hierarchical cluster analysis.
Pathways enrichment analysis
The same KEGG-IDs were used to analyze metabolic pathways using the Kyoto Encyclopedia of Genes and Genomes function (KEGG pathways)45,46,47 and Reactome pathway enrichment analysis yielded similar results about the specific pathways affected, allowing the expressions of proteins encoding enzymes found in the database to be mapped.
Conclusions
In verifying the differential expression of the cecal proteome in cattle, our results show that the blend of essential oils associated with α-amylase incorporated as a feed additive for beef cattle increased the expression of enzymes at dietary starch levels of 25%, 35%, and 45% compared with monensin. The higher expression of proteins related to carbohydrate degradation that participate in glycolysis and gluconeogenesis depended on increased feed intake and reduced protein synthesis expression. The optimum starch level was 35% for both feed additives; higher concentrations of starch (45%) increased the expression of inflammatory responses and reduced the expression of proteins involved in energy metabolism, probably due to damage to the cecum epithelium.
Data availability
The datasets used can be made available by the corresponding author on reasonable request.
References
-
Brake, D. W. & Swanson, K. C. Ruminant nutrition symposium: Effects of postruminal flows of protein and amino acids on small intestinal starch digestion in beef cattle. J. Anim. Sci. 96, 739–750 (2018).
-
Allen, M. S., Bradford, B. J. & Oba, M. Board-invited review: The hepatic oxidation theory of the control of feed intake and its application to ruminants. J. Anim. Sci. 87, 3317–3334 (2009).
-
Reynolds, C. K. & Maltby, S. A. Regulation of nutrient partitioning by visceral tissues in ruminants. J. Nutr. 124, 1399S-1403S (1994).
-
Lozano, O. et al. Net absorption and hepatic metabolism of glucose, L-lactate, and volatile fatty acids by steers fed diets containing sorghum grain processed as dry-rolled or steam-flaked at different densities. J. Anim. Sci. 78, 1364 (2000).
-
NASCEM. Nutrient Requirements of Beef Cattle 8th edn. (National Academies Press, 2016).
-
Silva, F. R. N. et al. Monensin residues in the production of Minas Frescal cheese: Stability, effects on fermentation, fate and physicochemical characteristics of the cheese. Food Res. Int. 137, 109440 (2020).
-
Meyer, N. F. et al. Effect of essential oils, tylosin, and monensin on finishing steer performance, carcass characteristics, liver abscesses, ruminal fermentation, and digestibility1. J. Anim. Sci. 87, 2346–2354 (2009).
-
Meschiatti, M. A. P. et al. Feeding the combination of essential oils and exogenous α-amylase increases performance and carcass production of finishing beef cattle1. J. Anim. Sci. 97, 456–471 (2019).
-
Toseti, L. B. et al. Effects of a blend of essential oils and exogenous α-amylase in diets containing different roughage sources for finishing beef cattle. Anim. Feed Sci. Technol. 269, 114643 (2020).
-
Harmon, D. L., Yamka, R. M. & Elam, N. A. Factors affecting intestinal starch digestion in ruminants: A review. Can. J. Anim. Sci. 84, 309–318 (2004).
-
Huntington, G. B., Harmon, D. L. & Richards, C. J. Sites, rates, and limits of starch digestion and glucose metabolism in growing cattle1. J. Anim. Sci. 84, E14–E24 (2006).
-
Moharrery, A., Larsen, M. & Weisbjerg, M. R. Starch digestion in the rumen, small intestine, and hind gut of dairy cows—A meta-analysis. Anim. Feed Sci. Technol. 192, 1–14 (2014).
-
Westreicher-Kristen, E. et al. Postruminal digestion of starch infused into the abomasum of heifers with or without exogenous amylase administration. J. Anim. Sci. 96, 1939–1951 (2018).
-
Owens, F. N., Zinn, R. A. & Kim, Y. K. Limits to starch digestion in the ruminant small intestine1,2. J. Anim. Sci. 63, 1634–1648 (1986).
-
Huntington, G. B. Starch utilization by ruminants: From basics to the bunk. J. Anim. Sci. 75, 852 (1997).
-
Lohrenz, A.-K. et al. Glucose transporters and enzymes related to glucose synthesis in small intestinal mucosa of mid-lactation dairy cows fed 2 levels of starch. J. Dairy Sci. 94, 4546–4555 (2011).
-
Mills, J. A. N. et al. A mechanistic model of small intestinal starch digestion and glucose uptake in the cow. J. Dairy Sci. 100, 4650–4670 (2017).
-
Pinto, A. C. J. & Millen, D. D. Nutritional recommendations and management practices adopted by feedlot cattle nutritionists: The 2016 Brazilian survey. Can. J. Anim. Sci. https://doi.org/10.1139/cjas-2018-0031 (2018).
-
Hoover, W. H. Digestion and absorption in the hindgut of ruminants. J. Anim. Sci. 46, 1789–1799 (1978).
-
Siciliano-Jones, J. & Murphy, M. R. Production of volatile fatty acids in the rumen and cecum-colon of steers as affected by forage: Concentrate and forage physical form. J. Dairy Sci. 72, 485–492 (1989).
-
Sanz-Fernandez, M. V. et al. Targeting the hindgut to improve health and performance in cattle. Anim. Open Access 10, 1817 (2020).
-
Choi, Y. J. et al. SERPINB1-mediated checkpoint of inflammatory caspase activation. Nat. Immunol. 20, 276–287 (2019).
-
Liu, J., Xu, T., Zhu, W. & Mao, S. High-grain feeding alters caecal bacterial microbiota composition and fermentation and results in caecal mucosal injury in goats. Br. J. Nutr. 112, 416–427 (2014).
-
Chang, G. et al. Sodium butyrate modulates mucosal inflammation injury mediated by GPR41/43 in the cecum of goats fed a high concentration diet. Front. Physiol. https://doi.org/10.3389/fphys.2019.01130 (2019).
-
Hristov, A. N., McAllister, T. A. & Cheng, K. J. Effect of dietary or abomasal supplementation of exogenous polysaccharide-degrading enzymes on rumen fermentation and nutrient digestibility. J. Anim. Sci. 76, 3146 (1998).
-
Thomas, M. et al. Metagenomic characterization of the effect of feed additives on the gut microbiome and antibiotic resistome of feedlot cattle. Sci. Rep. 7, 12257 (2017).
-
Steele, M. A., Penner, G. B., Chaucheyras-Durand, F. & Guan, L. L. Development and physiology of the rumen and the lower gut: Targets for improving gut health. J. Dairy Sci. 99, 4955–4966 (2016).
-
Tao, S. et al. A high-concentrate diet induced colonic epithelial barrier disruption is associated with the activating of cell apoptosis in lactating goats. BMC Vet. Res. 10, 235 (2014).
-
UniProt. Universal protein resource (Uniprot) (2020). http://www.uniprot.org/ (Accessed 1 April 2020).
-
Percie du Sert, N. et al. Reporting animal research: Explanation and elaboration for the ARRIVE guidelines 20. PLoS Biol. 18, e3000411 (2020).
-
Fox, D. et al. The Cornell net carbohydrate and protein system model for evaluating herd nutrition and nutrient excretion. Anim. Feed Sci. Technol. 112, 29–78 (2004).
-
McIntosh, F. M. et al. Effects of essential oils on ruminal microorganisms and their protein metabolism. Appl. Environ. Microbiol. 69, 5011–5014 (2003).
-
Association of Official Analytical Chemists (AOAC). Determination of Crude Protein in Animal Feed, Forage (Plant Tissues), Grain, and Oil Seeds Using Block Digestion with a Copper Catalyst and Steam Distillation into Boric Acid (AOAC International, 2001).
-
Van Soest, P. J., Robertson, J. B. & Lewis, A. B. Symposium: Carbohydrate methodology, metabolism, and nutritional implications in dairy cattle. J. Dairy Sci. 74, 3583–3597 (1991).
-
James, C. S. Analytical Chemistry of Foods (Blackie Academic & Professional, 1995).
-
Kononoff, P. J., Heinrichs, A. J. & Buckmaster, D. R. Modification of the Penn State Forage and total mixed ration particle separator and the effects of moisture content on its measurements. J. Dairy Sci. 86, 1858–1863 (2003).
-
Doumas, B. T., Bayse, D. D., Carter, R. J., Peters, T. & Schaffer, R. A candidate reference method for determination of total protein in serum. I. Development and validation. Clin. Chem. 27, 1642–1650 (1981).
-
Braga, C. P. et al. Mercury fractionation in dourada (Brachyplatystoma rousseauxii) of the Madeira River in Brazil using metalloproteomic strategies. Talanta 132, 239–244 (2015).
-
Santos, F. A. et al. Metallomic study on plasma samples from Nile tilapia using SR-XRF and GFAAS after separation by 2D PAGE: Initial results. Microchim. Acta 173, 43–49 (2011).
-
Neves, R. C. F. et al. Fracionamento de cobre em proteínas do plasma, músculo e fígado de tilápia do Nilo. Quim. Nova 35, 493–498 (2012).
-
Moraes, P. M. et al. A preliminary and qualitative metallomics study of mercury in the muscle of fish from Amazonas, Brazil. Biol. Trace Elem. Res. 150, 195–199 (2012).
-
Silva, F. A. et al. Selenium fractionation from plasma, muscle and liver of Nile tilapia (Oreochromis niloticus). J. Food Meas. Charact. 7, 158–165 (2013).
-
Shevchenko, A., Tomas, H., Havli, J., Olsen, J. V. & Mann, M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat. Protoc. 1, 2856–2860 (2006).
-
Conesa, A. et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005).
-
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 28, 27–30 (2000).
-
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
-
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M. & Tabane, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 49, D545–D551 (2021).
Acknowledgements
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brazil (CAPES)—Finance Code 001 and the authors thank the DSM/Tortuga nutritional products by financial support.
Author information
Authors and Affiliations
Contributions
L.R. designed and performed the experiment, analyzed samples and datas and wrote the manuscript. VC and AP designed and performed the experiment. M.B., J.A., C.B., A.A. and R.M. analyzed samples and datas. D.M., J.V. and P.P. designed, supervised and analyzed samples and datas. All authors helped to revise, read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
About this article
Cite this article
Rocha, L.C., Assunção, A.S.d.A., Martins, R.A. et al. Feedlot diets containing different starch levels and additives change the cecal proteome involved in cattle's energy metabolism and inflammatory response. Sci Rep 12, 5691 (2022). https://doi.org/10.1038/s41598-022-09715-7
-
Received:
-
Accepted:
-
Published:
-
DOI : https://doi.org/10.1038/s41598-022-09715-7
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.
campbellformiscrad.blogspot.com
Source: https://www.nature.com/articles/s41598-022-09715-7
0 Response to "Factorial Arrangement Feedlot Beef Steers Additives"
Postar um comentário